Can an AI agent help diagnose genetic diseases?
Examines whether LLM agents can handle a first-pass genomic analysis for pediatric rare disease cases. Lays out a three-agent workflow that maps symptoms to HPO terms, ranks candidate genes, and searches variants with purpose-built tools and prompts. Tests the system on Rett syndrome and Maple Syrup Urine Disease examples, where the agents successfully surface the causal variants. Emphasizes keeping human analysts in the loop and envisions agents revisiting unsolved cases as genomic knowledge grows. Describes the architectural choices that make the workflow extensible over time.
Introduction
Recently, I’ve been thinking about how advanced LLMs might be utilized in my own field, genomic medicine. While genomic medicine involves a lot of data processing, such as the execution of bioinformatics workflows to identify genetic variants from the output of DNA sequencing instruments, it also involves a lot of human effort. Genetic analysts must review a patient’s variants and attempt to identify a causal mutation given the patient’s symptoms. This requires specialized knowledge, experience, and software tools. The time investment of a human expert is often the limiting factor for organizations that provide genetic testing services, as advances in DNA sequencing technologies have made it possible to rapidly sequence far more patients than could be analyzed in a reasonable amount of time by even a large team of experts.
Could there be opportunities to delegate some of the work done by expert genetic analysts to LLMs? After all, lots of work is already delegated to software algorithms: DNA sequence alignment, variant detection, and variant annotation just to name a few.
In this post, I explore building an AI agent that could be used to autonomously perform a “first pass” analysis. The idea is to have the agent identify “low-hanging fruit” in genomic medicine: cases where the causative genetic variant can be identified by following a clear procedure. This might only represent a few percent of patients, but it could potentially save time and allow genetic analysts to focus more of their efforts on complex cases. As the capabilities of LLMs grow, an AI agent is likely to be able to handle increasingly complex cases over time, especially with genomic-specific training based on human-solved cases.
As with most uses of AI agents, it seems best to keep a human in the loop. This could be accomplished by having the agent present a report to a human genetic analyst for independent verification if it believes it identified a causative variant in the patient.
There is another potentially valuable use for these agents that I will discuss more at the end of this post: having autonomous agents look back at unsolved cases to see if updated scientific knowledge (such as newly characterized gene functions) allows for candidate variants to be identified that would have been overlooked in the original analysis. The diagnostic rate for pediatric rare diseases is only around 30%. As a result, the number of unsolved cases steadily grows. It is simply not possible for a team of human genetic analysts to revisit every unsolved case with any regularity, even though our knowledge of genes and diseases is constantly growing.
TL;DR
If you don’t want to read the entire post below, here are the key results:
I built a set of AI agents and associated tools tasked with following a minimal standard operating procedure for the identification of causative genetic variants for pediatric rare diseases. I tested the agent with two artificially-generated scenarios:
- A patient whose symptoms described Rett syndrome and had a ClinVar-characterized heterozygous deleterious variant in the MECP2 gene.
- A patient whose symptoms described Maple Syrup Urine Disease and had an uncharacterized homozygous deleterious variant in the BCKDHA gene.
In both scenarios, the agent successfully identified the causative variant out of the full exome sequencing results. Continue reading for additional details on both the design of the agents and the two scenarios.
Analysis Workflow
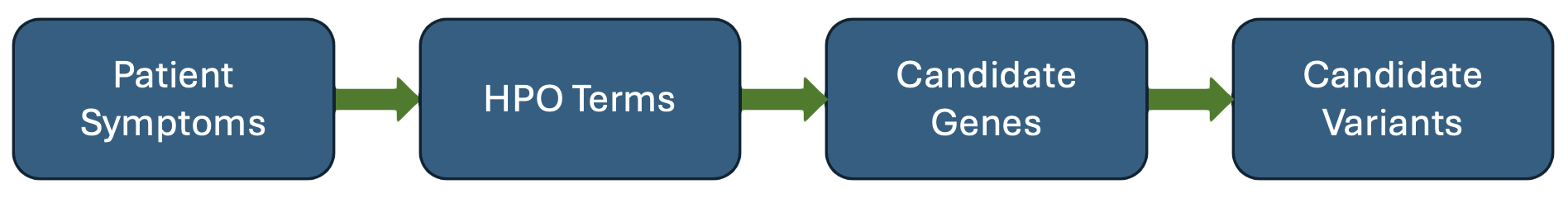
For rare pediatric genetic diseases, which will be the focus of my agent, a standard analysis workflow might look like this:
- Obtain a description of the patient’s symptoms
- Match the symptoms to a controlled vocabulary of phenotypes, such as Human Phenotype Ontology (HPO) terms
- Use existing phenotype-to-gene databases to identify candidate genes with known links to the patient’s HPO terms
- Search the patient’s genomic data for deleterious variants in the candidate genes, prioritizing genes that could explain the greatest number of the patient’s symptoms
- Use population databases, such as gnomAD, to filter out variants that occur at relatively high frequency in the general population
This workflow will identify potentially diagnostic variants in many well-characterized diseases that can be caused by the disruption of a single gene (called monogenic diseases).
Agent Architecture
State-of-the-art LLMs have a number of features that allow them to be applied to a range of tasks:
- They’ve been trained on vast amounts of human knowledge, including genetics
- They are quite good (but not perfect) at following instructions
- They have some facsimile of human reasoning that allows them to take a complex task and break it down into manageable chunks
- They can be given access to specialized tools, and they will use those tools to complete a given task
OpenAI has an Agents SDK that allows one to harness these features to build custom agents. I decided to use their SDK to fully implement the analysis workflow described above. The approach I took breaks the workflow down into three sequential components, each implemented as a separate agent:
- Start with a description of the patient’s symptoms
- Determine HPO terms from the description
- Identify candidate genes from the list of HPO terms
- Identify candidate variants based on the list of candidate genes
I plan to write follow-up posts with a deep-dive into the implementation of each agent. For now, I’ll briefly describe each one and the tools I provide it to accomplish its task. The critical components of an agent include the following:
- Instructions or “prompts”: This is where the agent’s task must be clearly defined. A description of what input will be provided to the agent, the tools it has access to for obtaining additional information, and the desired output should all be specified in the instructions.
- Tools: These are software programs that the agent can use to obtain additional information. It can be a generic tool, like the ability to perform a web search, or a custom tool written specifically for the agent to use. In the OpenAI Agents SDK, any Python function can be presented to an agent as a tool.
- Input: This is the information provided to the agent. Depending on the LLM used, it could be text, images, or audio.
- Output: Defining the structure of the output of an agent can be critically important, as the output might need to be used as input for another agent, or consumed by another system that requires a specific data format.
The HPO Agent
The job of the HPO Agent is to take a text description of the patient’s phenotype and turn it into a list of HPO terms. To assist the agent, I wrote an hpo_search tool in Python that performs a vector similarity search to retrieve the “nearest” HPO terms for a given query. The agent’s task is to identify the individual symptoms in the provided description, search for each one using the hpo_search tool, and build a list of relevant HPO terms from the results.
The Gene Agent
The Gene Agent runs after the HPO Agent, and is provided the list of HPO terms. It has access to another simple Python tool I wrote that looks up associated genes for a list of HPO terms using a downloaded phenotype-to-genes data file from the HPO website. The tool ranks the genes by counting how many of the provided HPO terms are associated with each gene. The agent is instructed to output a prioritized list of genes based on the tool’s results.
The Variant Agent
Finally, we have the Variant Agent. With the prioritized list of genes, this agent is tasked with querying the patient’s genomic sequencing results to locate candidate variants that can potentially explain the patient’s symptoms. To do this, I wrote a query tool for the agent to use. The tool searches a database table of annotated variants imported from the patient’s full sequencing results. The tool supports four filters:
- Gene: Restricts the returned variants to those within a specific gene.
- ClinVar Classification: ClinVar is a public database of observed human genetic variants and their roles in disease. As organizations all over the world contribute data to ClinVar, it’s a critical resource to consider in genomic medicine.
- Variant Consequence: Annotation tools assign a consequence to variants found within a gene, allowing for some inference as to the effect of the variant on the gene’s function.
- gnomAD Frequency: The variant’s observed frequency in the population database gnomAD. For rare diseases, the causative variants are almost always very rare in the human population, otherwise the disease itself wouldn’t be rare.
The instructions for the Variant Agent provide guidelines on using the variant query tool, including the possible values for the ClinVar and Variant Consequence filters. It also suggests using a gnomAD allele frequency maximum of 0.5%, an appropriate value for pediatric rare diseases.
The variant query tool outputs a summary of each matching variant that looks like this:
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
<variant>
Chromosome: chr7
Position: 147395769
Ref Allele: G
Alt Allele: A
Gene Symbols: CNTNAP2, gnomAD AF: 0.171863
ClinVar Summary: ClinVar ID: VCV000095556, Reference allele: G, Alternate allele: A, Allele-specific: True, Classification: Benign, Review Status: criteria provided, multiple submitters, no conflicts, Diseases: Pitt-Hopkins-like syndrome, Inborn genetic diseases, not specified, not provided, Cortical dysplasia-focal epilepsy syndrome
HGNC: CNTNAP2, transcript: ENST00000361727.7, consequences: synonymous_variant, hgvsc: ENST00000361727.7:c.1659G>A
HGNC: CNTNAP2, transcript: NM_014141.5, consequences: synonymous_variant, hgvsc: NM_014141.5:c.1659G>A
HGNC: CNTNAP2, transcript: ENST00000637694.1, consequences: non_coding_transcript_exon_variant, hgvsc: ENST00000637694.1:n.1562G>A
HGNC: CNTNAP2, transcript: ENST00000638117.1, consequences: non_coding_transcript_exon_variant, hgvsc: ENST00000638117.1:n.1562G>A
HGNC: CNTNAP2, transcript: ENST00000636870.1, consequences: non_coding_transcript_exon_variant, hgvsc: ENST00000636870.1:n.1521G>A
HGNC: CNTNAP2, transcript: ENST00000637825.1, consequences: non_coding_transcript_exon_variant, hgvsc: ENST00000637825.1:n.1142G>A
Genotype: 0/1, GQ: 42.0
Total Depth: 47, Allele Depths: [29, 18]
Maternal Genotype: 0/1, Paternal Genotype: 0/1
Variant Type: SNV
</variant>
With the above instructions and access to the variant query tool, the agent should go through the prioritized list of genes and attempt to locate candidate variants using appropriate search criteria.
Does it work?
Test setup
To test whether these agents can identify a potential causative variant given both a description of a patient’s symptoms as well as access to their annotated genomic sequencing results, I needed to gather a few things:
- A realistic description of symptoms matching a rare pediatric disease
- A causative genetic variant
- Genomic sequencing data (either exome sequencing or whole genome sequencing)
For the symptom description, I asked ChatGPT o4-mini-high to provide an example phenotype description for a pediatric rare disease. It selected Rett syndrome and provided this patient description:
The patient is an 18-month-old female. The patient was born at term after an uncomplicated pregnancy and delivery. Early development appeared normal: she held her head steadily by 3 months, rolled over by 5 months, and sat unsupported at 7 months. She began babbling at 6 months and used single consonant–vowel syllables (“ma,” “ba”) by 9 months. Starting around 12 months of age, her parents noted a gradual loss of previously acquired purposeful hand skills and spoken sounds. Over the next 6 months she stopped babbling altogether and developed repetitive hand-wringing movements, breath-holding spells, and episodes of hyperventilation.
It also mentioned that the disease is caused by a mutation in the MECP2 gene. To select a causative variant, I searched ClinVar for MECP2 and selected a pathogenic missense variant.
The last component needed to test the agents is a person’s exome or whole-genome sequencing results into which we can insert the MECP2 variant. For this, I decided to use my own personal DNA sequencing, which I had performed by a commercial company back in 2021. I’ll go into more detail in a later post, but the overall process was:
- Start with the variants from my DNA sequencing in Variant Call Format (VCF)
- Add the pathogenic MECP2 variant from ClinVar
- Annotate the variants with Illumina’s Nirvana variant annotation tool, which will add (among other annotations):
- gnomAD frequency
- ClinVar classifications
- Variant consequences
- Import the resulting annotated variants into a database table to enable rapid querying (there are about 48,000 variants after filtering down to an exome equivalent)
- Since I’m male and the test scenario refers to a female patient, I randomly modified my X chromosome variants to be either heterozygous or homozygous instead of hemizygous.
- Genetic testing often involves sequencing the patient as well as both parents when possible. As I only have my own sequencing, I added paternal and maternal genotypes as heterozygous to the database, except for the MECP2 variant, where I kept the parental genotypes as reference to simulate a de novo variant in the patient.
Running the agents
With all the above setup complete, I ran the agents. First up is the HPO Agent:
Input: The patient is an 18-month-old female. The patient was born at term after an uncomplicated pregnancy and delivery. Early development appeared normal: she held her head steadily by 3 months, rolled over by 5 months, and sat unsupported at 7 months. She began babbling at 6 months and used single consonant–vowel syllables (“ma,” “ba”) by 9 months. Starting around 12 months of age, her parents noted a gradual loss of previously acquired purposeful hand skills and spoken sounds. Over the next 6 months she stopped babbling altogether and developed repetitive hand-wringing movements, breath-holding spells, and episodes of hyperventilation.
- HP:0032588: Hand apraxia
- Reasoning: This term best fits the description of ‘loss of purposeful hand skills’ as it specifically refers to the inability to perform learned hand movements, with a high similarity score (0.534).
- HP:0007064: Progressive language deterioration
- Reasoning: This accurately reflects the ‘gradual loss of previously acquired purposeful hand skills and spoken sounds,’ specifically regarding language, as described in the case. It is the highest-scoring, most specific match (0.632).
- HP:0012171: Stereotypical hand wringing
- Reasoning: The patient developed ‘repetitive hand-wringing movements’; this term directly matches the description with a strong similarity score (0.666).
- HP:6000950: Breath-holding spell
- Reasoning: The description of ‘breath-holding spells’ is specifically captured by this term, which refers to episodic breath-holding in children. It is accurate and highly relevant (0.558).
- HP:0002883: Hyperventilation
- Reasoning: The patient’s ‘episodes of hyperventilation’ are directly described by this HPO term, which was the top and most specific result (0.659).
Using the search tool, the agent performed individual searches for each symptom and returned 5 HPO terms, along with its reasoning for each selection.
Next was the Gene Agent, which was provided the 5 HPO terms above:
- Gene: MECP2, Rank: 1
- Reasoning: MECP2 is associated with 4 of the 5 provided HPO terms—hand apraxia, progressive language deterioration, stereotypical hand wringing, and hyperventilation. This pattern of symptoms strongly overlaps with the classical presentation of Rett syndrome, for which MECP2 is the primary gene. This makes MECP2 the top-ranked gene.
- Gene: GABBR2, Rank: 2
- Reasoning: GABBR2 is associated with three key HPO terms: hand apraxia, stereotypical hand wringing, and hyperventilation. This gene has recently been linked to Rett-like phenotypes and shares much of the clinical spectrum of MECP2-related disorders.
- Gene: CNTNAP2, Rank: 3
- Reasoning: CNTNAP2 is associated with progressive language deterioration, stereotypical hand wringing, and hyperventilation, overlapping with much of the neurodevelopmental phenotype in the patient. Mutations in CNTNAP2 are linked to neurodevelopmental disorders with regression, and stereotypies.
- Gene: CDKL5, Rank: 4
- Reasoning: CDKL5 is associated with 3 of the key terms: hand apraxia, stereotypical hand wringing, and hyperventilation. CDKL5 mutations cause early-onset epileptic encephalopathy with features overlapping classic Rett syndrome, including the symptoms described in the case.
- Gene: CASK, Rank: 5
- Reasoning: CASK overlaps with stereotypical hand wringing and hyperventilation. Mutations in this gene can cause symptoms overlapping Rett syndrome.
- Gene: SMC1A, Rank: 6
- Reasoning: SMC1A is associated with both hand apraxia and stereotypical hand wringing. Mutations here can present with neurodevelopmental disorders including stereotypies.
The previous agent’s HPO selection appeared to be quite effective, as the MECP2 gene was ranked first by the gene agent! With the ranked set of genes, I finally ran the Variant Agent:
I identified a de novo, pathogenic MECP2 missense variant that fully explains the patient’s clinical presentation.
- MECP2 (X-linked; Rett syndrome)
• Genomic coordinate (GRCh38): X:154,030,863 G>A
• Transcript: NM_004992.3:c.965C>T (p.Thr322Met)
• Consequence: missense_variant
• gnomAD v4.0 AF: 0.0 (absent)
• ClinVar: Pathogenic (VCV000143754), reviewed by expert panel; associated with Rett syndrome
• Patient genotype: 0/1 (heterozygous)
• Mother: 0/0 (homozygous reference)
• Father: 0 (hemizygous reference)
• Inheritance: confirmed de novo in the patientClinical correlation: De novo pathogenic MECP2 variants are the established cause of classic Rett syndrome, which presents in females with normal early development followed by regression of language and hand skills, stereotyped hand-wringing, and breathing irregularities. This variant fully accounts for the patient’s phenotype.
Success! The variant X:154,030,863 G>A is the exact one I inserted based on the ClinVar record. Since I modified the parental genotypes to be reference, the agent correctly reported it as a de novo variant.
You can see from the agent’s output above that it first searched the top-ranked gene MECP2 for a set of consequences that have a high chance of being deleterious: stop_gained, frameshift_variant, splice_acceptor_variant, splice_donor_variant, missense_variant, start_lost. It included a gnomAD frequency maximum of 0.005 (0.5%), as suggested in the agent instructions.
A second test
I decided to try another test, this time creating a novel variant that would not be classified in ClinVar but would be deleterious to a gene implicated in a rare disease. I also wanted to try an example of an autosomal recessive disease. Again, I asked ChatGPT o4-mini-high to generate a scenario:
The patient is a 6-day-old male. The patient was born at 39 weeks after an uncomplicated pregnancy. Birth weight was 3.2 kg and Apgars were 8 and 9 at 1 and 5 minutes. Newborn screening at 48 hours reported markedly elevated branched-chain amino acids. On day 4 of life he developed poor feeding and repeated emesis. By day 5 the infant exhibited lethargy and intermittent episodes of tachypnea. Parents noted a “sweet, burnt-sugar” odor in the diapers.
ChatGPT indicated these symptoms could be caused by mutations in both copies of BCKDHA, so I defined a homozygous frameshift mutation early in the gene’s coding sequence, annotated it, and inserted into the database used by the variant query tool alongside my own variants.
I then ran all three agents in sequence, with the output shown below. I’ve truncated the ranked gene list after 5 genes for space reasons, but the full output is available.
- HP:0008344: Elevated circulating branched chain amino acid concentration
- Reasoning: This term was selected because newborn screening showed markedly elevated branched-chain amino acids in the blood, and this was the top-matching, clinically precise term referring to blood levels.
- HP:0008872: Feeding difficulties in infancy
- Reasoning: Poor feeding was described on day 4 of life. This term is clinically appropriate as it specifies feeding difficulty in infancy with the highest similarity score among infant-specific terms.
- HP:0001254: Lethargy
- Reasoning: By day 5 the infant was described as lethargic, and this matches the term ‘Lethargy’ directly, with a high similarity score and clinical specificity.
- HP:0033072: Abnormal macroscopic urine appearance
- Reasoning: A ‘sweet, burnt-sugar’ odor in the diapers was noted, which is reflected by this term’s inclusion of abnormal urine odor.
- Gene: BCKDHA, Rank: 1
- Reasoning: BCKDHA is associated with three of the submitted HPO terms: elevated circulating branched chain amino acid concentration (HP:0008344), feeding difficulties in infancy (HP:0008872), and lethargy (HP:0001254). Importantly, BCKDHA mutations cause Maple Syrup Urine Disease (MSUD), which is strongly associated with all the above features and the classic abnormal urine odor. The gene directly fits the clinical scenario of abnormal branched-chain amino acids, lethargy, feeding problems, and typically abnormal urine.
- Gene: CPT2, Rank: 2
- Reasoning: CPT2 is associated with lethargy (HP:0001254), abnormal macroscopic urine appearance (HP:0033072), and feeding difficulties in infancy (HP:0008872). While CPT2 deficiency can present with lethargy and feeding issues, abnormal branched chain amino acids are not characteristic for CPT2. The gene is included due to HPO matches, but does not explain the key metabolic laboratory finding as well as BCKDHA.
- Gene: KCNA1, Rank: 3
- Reasoning: KCNA1 is linked to feeding difficulties (HP:0008872) and lethargy (HP:0001254). However, it is not associated with abnormal branched chain amino acids or abnormal urine appearance, making it less likely given the submitted clinical scenario.
- Gene: PLCH1, Rank: 4
- Reasoning: PLCH1 is associated with feeding difficulties and lethargy but not with branched chain aminoacidemia or abnormal urine appearance. Less likely than genes affecting metabolism directly.
- Gene: GATA1, Rank: 5
- Reasoning: GATA1 is matched to lethargy and abnormal urine, without a direct correlation with branched chain amino acids or classic metabolic presentations such as MSUD.
We identified a single, rare, biallelic‐inactivating variant in BCKDHA that is diagnostic for autosomal recessive Maple Syrup Urine Disease Type 1A.
1) BCKDHA NM_000709.3:c.5del
– Consequence: frameshift_variant (p.(Val2Trpfs*?) )
– gnomAD v4.0 allele frequency: 0.0
– ClinVar: not previously reported
– Patient genotype: 1/1 (homozygous alternate; total depth 19×, allele depths 0/19)
– Maternal genotype: 0/1 (heterozygous carrier)
– Paternal genotype: 0/1 (heterozygous carrier)Interpretation
- The patient is homozygous for a novel frameshift deletion near the N-terminus of BCKDHA, predicted to abolish normal protein function.
- Both parents are heterozygous carriers, consistent with autosomal recessive inheritance.
- Loss‐of‐function variants in BCKDHA are a well‐established cause of Maple Syrup Urine Disease, matching the patient’s elevated branched‐chain amino acids, feeding difficulties, lethargy and “sweet, burnt‐sugar” odor.
Conclusion
This homozygous BCKDHA frameshift variant provides a molecular diagnosis of Maple Syrup Urine Disease (MSUD) Type 1A. Given the low genotype quality (GQ 4) for the proband, we recommend confirmation by orthogonal testing (e.g., Sanger sequencing) and prompt initiation or continuation of appropriate metabolic management.
Again the agent was able to rank the relevant gene BCKDHA as #1 due to the gene’s known involvement in most of the identified phenotypes. By querying for rare variants in that gene, it was able to locate the causative variant that I added by specifying appropriate consequence terms.
Limitations and challenges
While the above two examples were successfully analyzed by the agents, it’s important to keep in mind that these were straightforward cases involving well-characterized diseases with “obvious” causative variants that a genetic analyst would rapidly have identified.
However, even this limited capability can be useful, especially as a tool for revisiting unsolved cases to see if new knowledge can point to a causative variant that was previously overlooked. Imagine agents running regularly in the background, periodically re-analyzing unsolved cases. They would alert genetic analysts only when a potential diagnostic variant is identified. This could put at least a small dent in the accumulating number of unsolved cases as the expansion of genetic testing continues, with minimal burden on genetic analysts. Nevertheless, serious concerns remain about AI safety, reliability, and interpretability. We must avoid risks like hallucinated symptoms or variants not present in the data, as well as guard against prompt injection attacks that cause unintended behaviors.
I believe it is quite clear that advanced LLMs, given high quality instructions and access to appropriate tools, can play a role in genomic medicine. Given their ability to integrate information and use tools, they may even succeed at more complex cases. I’d like to try the agent on slightly more difficult cases - perhaps a situation where two diagnostic variants need to be identified, as with compound heterozygosity. By testing the agents on a variety of cases, we can identify their limitations and explore improvements, such as refining instructions or providing new tools.
A significant limitation of the minimal approach I implemented is its over-reliance on only three variant annotations: ClinVar classifications, gnomAD frequency, and transcript consequences. While these are highly useful, there are many other annotations and tools that the agent could utilize when analyzing variants. For example, a very new tool from Google, AlphaGenome, produces a wide variety of functional effect predictions for variants, including tissue-specific effects, that could be rapidly analyzed by an agent to determine if a suspect variant is likely to play a role in the patient’s symptoms.
Another critical resource for human genetic analysts is the medical research literature. Often, an analyst will find a rare variant that disrupts a gene whose function is not well understood. A literature search might reveal recent research connecting the gene to biological processes or pathways that might be relevant to the patient’s symptoms. OpenAI already provides a web search tool that can be made available to agents, allowing for access to publicly-available scientific literature. A custom tool designed specifically for querying medical research publications might be highly effective when provided to an agent tasked with investigating a gene of unknown function.
As you can see, there are a lot of potential paths of exploration for utilizing AI agents in genomic medicine. As I mentioned in the introduction, however, it’s important to treat these AI agents as specialized tools that can gather and summarize information, ultimately reporting back to human experts for final review.
What’s next
I plan to write individual posts describing how I implemented each of the three agents, including the prompts and tools I developed. These will be more technically focused than this post, which I wrote more to demonstrate the exciting potential of AI agents in the field.
I also hope to try out more challenging cases - perhaps by simulating various case reports described in the literature that had unique challenges.
In addition, I’m excited to see what novel uses of LLMs the field comes up with - whether that be autonomous agents running in the background examining unsolved cases, or research assistants that work alongside genetic analysts, rapidly summarizing new information about a poorly-characterized gene or variant.